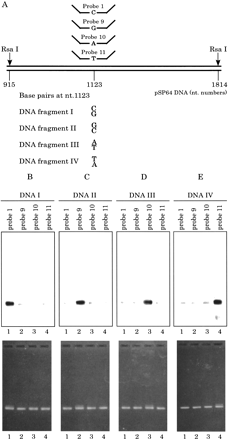
Fidelity of the marking (probing). A DNA fragment (900 bp) of pSP64 DNA with a different bases at position nt. number 1123 was subjected to direct marking using four different probing oligonucleotides (20-bp complimentary bases) and a splint oligonucleotide (17 bp). (A) A diagrammatic representation of probing oligonucleotides and target DNA fragments used. One mismatched base is present at position 10 of the 20 complementary base sequences in the probing oligonucleotides. DNA fragment I to IV represented pSP64 DNA with an altered base at position nt. number 1123, respectively. (B) Direct marking for a DNA fragment I. (Lanes 1–4) Probes 1, 9, 10, and 11 were used, respectively. (C) Direct marking for a DNA fragment II. (Lanes 1,–4) Probes 1, 9, 10, and 11 were used, respectively. (D) Direct marking for a DNA fragment III. (Lanes 1–4) Probes 1, 9, 10, and 11 were used, respectively. (E) Direct marking for a DNA fragment IV. (Lanes 1–4) probes 1, 9, 10, and 11 were used, respectively. The construction of the target DNA fragments is described in the Methods. (B–E, top) autoradiographic pattern; (bottom) ethidium bromide staining pattern. Nucleotide numbers (nt. numbers) are those registered in GenBank (accession no. X65328). For details, see Methods.











