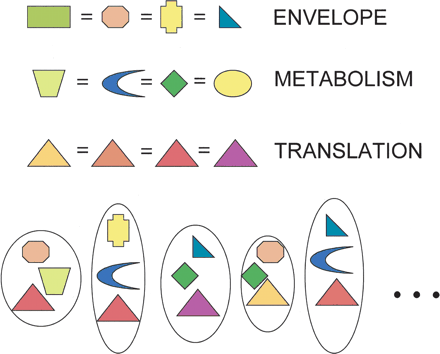
A mix-and-match model for prokaryotic genome evolution. Every cell needs genes for multiple functions, and new genomic lineages arise in evolution through mixing and matching of genes performing these different functions, by processes of replacement, including nonorthologous displacement (Koonin et al. 1996). The simplest hypothesis would be that all functions are equally subject to such exchange processes. For many functions, available genes include nonhomologs and even null entries (gene and function loss), indicated here by different shapes. Thus, for these functions, no genes or even gene families will likely appear to be shared among all genomes. For some informational functions especially (such as translation), displacement most often involves genes that, although evolutionarily distinct (as indicated by colors), are homologous (as shown by shape). Such genes will appear among those of the ubiquitous core.











