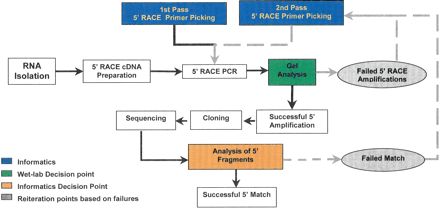
Overview of high-throughput workflow. The initiation of this workflow begins with the selection of genes for experimental verification. The gene models associated with these genes are used to select 5′ RACE–PCR primers (1st pass) (for more details, see Supplemental information). The primers are used to amplify RACE templates that have been generated in parallel. Failed amplifications are identified via agarose gel analysis, and new primers are picked for those genes (2nd pass). The products of the successful amplification are ligated into a plasmid vector. Eight clones from every amplification are sequenced and analyzed by comparison to the existing gene model. Amplified fragments that do not match their associated gene model are evaluated, and new primers are picked for the associated gene model (2nd pass). Successful matches are further evaluated in terms of whether they modify the current gene model, and the sequence is submitted to GenBank.











