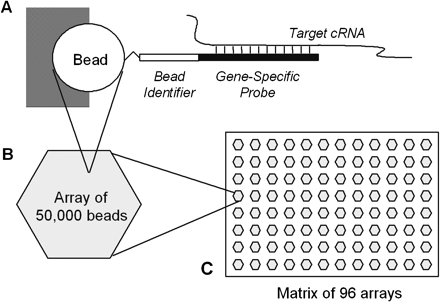
Design of a randomly assembled gene-specific probe array. (A) Representation of an individual bead lodged in a well. Attached to the bead by its 5′ end is a chimeric oligonucleotide ∼75 nucleotides in length, comprising an ∼25-nucleotide identifier sequence and a 50-nucleotide gene-specific probe. The bead identifier sequence is decoded using an algorithm described previously (Gunderson et al. 2004). We tested gene-specific probes of 25 and 50 bp in length and found that the 50 mers showed superior performance, consistent with prior findings (Hughes et al. 2001). The drawing is not to scale; the relative size of the oligonucleotide has been vastly exaggerated to show its features. (B) There are ∼50,000 beads in an ∼1.4-mm diameter optical fiber bundle, each bead lodged in a well at the end of an individual fiber in the bundle. (C) The bundles are arranged in a 96-array matrix matching the format of a standard microtiter plate.











