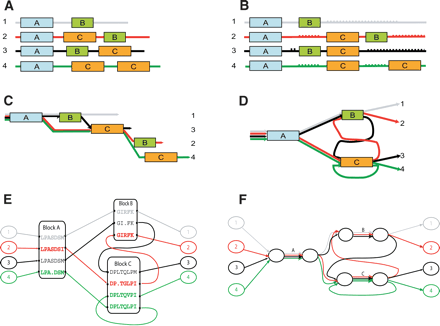
(A) Three linear paths represent three sequences: S1, S2, and S3. Vertices represent positions in each sequence and are labeled by the amino acid at that position. Vertices corresponding to aligned positions in the set of pairwise alignments are joined by dark edges. These edges give a set of “gluing” instructions, and we obtain the A-Bruijn graph by collapsing each set of glued vertices into a single vertex. (B) The resulting A-Bruijn graph contains a short (undirected) cycle on vertices I, R, and F. Edge labels indicate the multiplicity of the edge. (C) We remove short cycles and collapse each simple chain into a single edge, obtaining a simplified A-Bruijn graph. Each multiple edge has a label of the form l(m), where l is the length of the sequences represented by that edge and m is the multiplicity of the edge.











