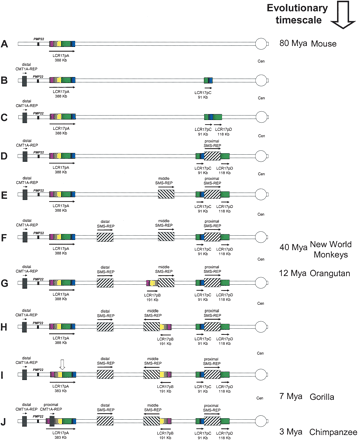
Rearrangements of proximal 17p LCR during primate evolution. Proximal chromosome 17p is depicted by two thin horizontal lines with the centromere (○) shown to the right. LCRs are shown as horizontal rectangles with the same color or black-and-white graphic, representing highly homologous sequence. (A) In the mouse genome, only LCR17pA is present. (B,C) The proximal portion of LCR17pA was duplicated >25 Mya. Note that LCR17pC and LCR17pD represent two overlapping portions of LCR17pA. (D) The proximal SMS-REP split the LCR17pC and LCR17pD copies resulting in three directly adjacent large LCRs. Two tandem duplications resulted in directly oriented middle (E) and distal (F) SMS-REPs (Park et al. 2002). (G) After the divergence of orangutan and gorilla 7–12 Mya, the distal portion of LCR17p was tandemly duplicated creating a directly oriented LCR17pB copy. (H) Both middle SMS-REP and LCR17pB, adjacent to it, were inverted. (I) Following that, at the junction between LCR17pA/B and LCR17pD copies, the evolutionary translocation t(4;19) occurred in a pre-gorilla individual, 7–12 Mya (vertical open arrow). (J) Lastly, between gorilla and chimpanzee, 3–7 Mya the proximal CMT1A-REP, present only in human and chimpanzee, resulted from the insertional duplication of the distal copy (Kiyosawa and Chance 1996; Reiter et al. 1997; Inoue et al. 2001). (Right) Time line of mammalian, mainly primate, evolution with million of years (Mya), as indicated.











