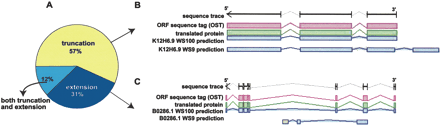
Cloning success based on the nature of repredictions. (A) Of ORFs cloned in ORFeome Version 3, 57% were repredicted to be shorter and 31% to be extended at one or both ends, whereas 12% of the cloned ORFs have been extended at one end and truncated at the other end. (B) Example of an ORF that was successfully cloned in ORFeome Version 3 after having been truncated at the 3′-end. The exon/intron structures in blue represent the old (K12H6.9WS9) and new (K12H6.9WS100) predictions of K12H6.9. Using primers based on WS100 and sequencing the resulting PCR product, we obtained a sequence trace (black arrow) that aligned to the WS100 prediction, showing a full-length OST (pink) of the exact structure predicted. The translated protein is shown in green, demonstrating that the cloned ORF is, indeed, in-frame. The primer designed for the 3′-end of the WS9 prediction cannot anneal to the coding sequence of the WS100 prediction explaining earlier cloning failure. (C) Example of an ORF that was successfully cloned in ORFeome Version 3 after having been extended at both ends. The 5′-primer based on WS9 is annealing in the middle of an intron in the new predicted gene model, explaining earlier cloning failure.











