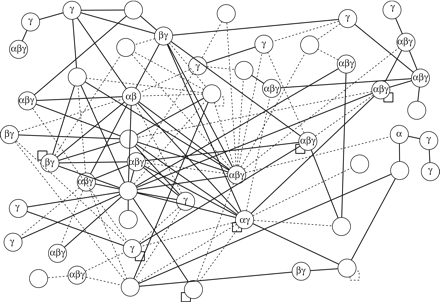
Protein interaction map of Kaposi's sarcoma associated virus (KSHV), highlighting conserved interactions and data quality. The protein interaction map of KSHV involves roughly half of all KSHV proteins. We assume that the other proteins interact with host proteins or their interactions have not yet been identified. Protein conservation is indicated by Greek letters, namely that the protein is conserved in α herpesviruses (HSV1), β herpesviruses (CMV), and/or γ herpesviruses (EBV). Solid lines indicate interactions that have been confirmed by coimmunoprecipitation; hatched lines were found by two-hybrid only. Note that only three of these interactions have been published thus far; the remaining ones are derived from a recent genome-wide two-hybrid screen (from P. Uetz, Y.A. Dong, C. Zeretzke, M. Roupelieva, D. Rose, C. Atzler, and J. Haas, in prep.). Interestingly, the proteins in this map are much better connected than in the T7 map (Fig. 2), and interactions of conserved proteins appear to be more reproducible than those of KSHV-specific proteins.











