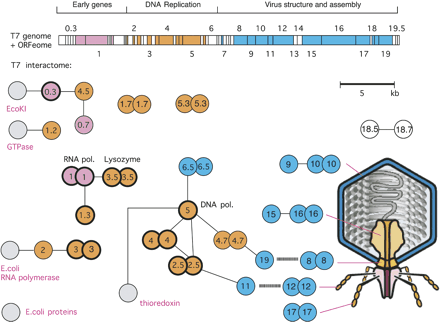
ORFeome and protein interaction map in bacteriophage T7. Cloned ORFs and random fragments were used to generate a two-dimensional interaction map. ORFeomes can also be used by structural genomics projects to derive 3D structures. A combination of interaction maps and crystal structures often allows the reconstruction of protein complexes as in viral particles or their subunits. The top panel shows the T7 genome with each ORF represented as a box. Note that only the main ORFs are indicated by integral numbers as originally identified by genetic screens. Genes in between have decimal numbers. Proteins that have been found to interact with others are indicated by colored boxes; box colors indicate a simplified assignment to functional classes as shown by the heading. Interactions between proteins are shown as lines, with self-interactions shown as dimers. Hatched lines indicate expected interactions which have not been found by two-hybrid analysis. Gray proteins correspond to host proteins (E. coli). Blue proteins are involved in virus assembly and structure, and their rough location in the virus particle is shown on the right, if known. Proteins with thick borders have been crystalized and their structure is known. Genetic map modified after Dunn and Studier (1983). Interactions primarily based on data from Bartel et al. (1996).











