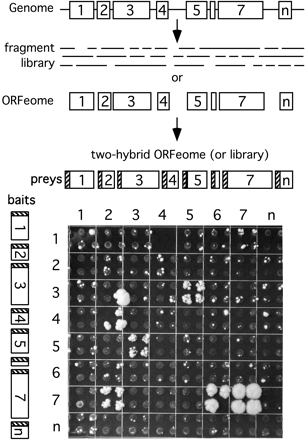
From viral ORFeome to protein interaction map. ORFs can be predicted from sequenced genomes with high confidence and subsequently cloned. Alternatively, libraries of random fragments can be generated. ORFs or fragments can then be cloned into two-hybrid vectors and tested for pairwise interactions in an array format (Uetz 2002). In this experiment, ORFs from KSHV were tested for interactions. Note that each test was done in quadruplicate to ensure reproducibility. Interactions between bait 3 and prey 5 (3-5) is also reproducible in the inverted configuration (bait 5 and prey 3), whereas interactions 4-2 and 7-2, and 7-6 are not. ORF 7 is homodimerizing (7-7). Colony 3-2 is a clear false positive as it is not reproducible in quadruplicate; 4-2 and 7-6 are considered true positives, although only three of four colonies are growing, which may be due to imprecise spotting. Also shown is the noticeable background growth, which can be easily distinguished from the two-hybrid signal in this array format.











