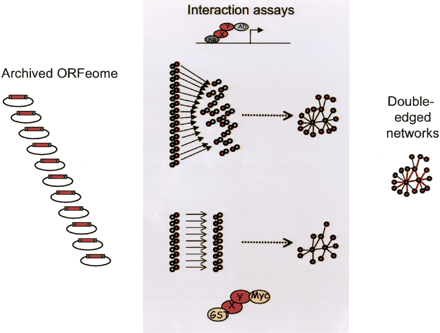
Modeling cellular networks requires efficient recombinational cloning “operating systems.” (A) Physical interactions. This example illustrates how the mapping of physical interactions between proteins can benefit from the use of two different assays (“Double-edged” networks). Here the yeast two-hybrid (Y2H) system is first used with individual DB-X baits and a pooled AD-ORFeome library (Reboul et al. 2003). Subsequently, positive Y2H interactions are retested using a different assay (Li et al. 2004): GST pull-down followed by anti-Myc Western-blot analysis. To express high numbers of proteins with the appropriate tags (DB, DNA binding domain; AD, activation domain; GST, glutathione-S-transferase), large collections of archived protein encoding open reading frames (ORFeomes) need to be available (efficiency), in ways that allow their subcloning in many different vectors (adaptability). The resulting “double-edged” networks are of greater overall quality.











