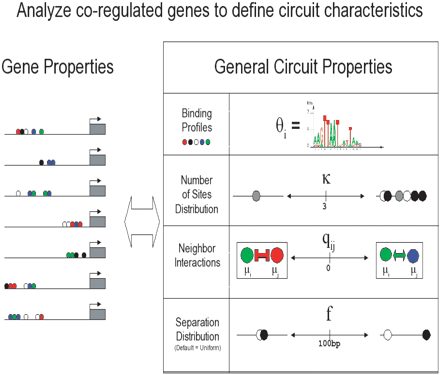
General parameters that the module sampler attempts to discover. A priori, motif binding models were modeled by uniform Dirichlet prior models. Nearest neighbor interactions were modeled as transition probabilities of a Markov chain. This allows us to calculate the posterior mean estimate of the transition probabilities based on the number of times that each specific type of binding site follows another and prior pseudocounts. A priori, we assume that all neighboring pairs also have uniform Dirichlet prior models. The algorithm also allows us to draw inferences regarding the number of sites per sequence. We chose a prior distribution on the number of sites per sequence based upon the distribution of reported sites. The separation distance was modeled as a flat function truncated at 100 bp.











