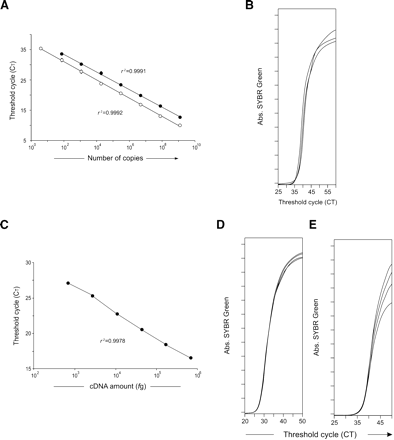
Linearity of double-round PCR amplification. (A) A double-stranded synthesized DNA sequence from Gzma was amplified in quadruplicate by a single PCR (•) or by a two-step PCR of 15 preamplification cycles (○). Decreasing template concentrations by a factor of 16 were used. Means ± SD of quadruplicates are included, although SDs frequently overlap with the symbols as we found little or no variation. Linear regression curves of these amplified standards are indicated and have similar high correlation coefficients (r2 = 0.999). Similar results were obtained with three different types of synthesized cDNA. (B) Four molecules of a double-stranded synthesized Gzma sequence were amplified in triplicate using 15 cycles on the first PCR amplification followed by a second real-time PCR. PCR accumulation curves are shown. (C) A bulk cDNA population (7 × 105 to 680 fg) was used to amplify 28S (Mrp-S21) gene in quadruplicate by a two-step PCR of 15 preamplification cycles. Decreasing template concentrations by a factor of 4 were used. Mean values and standard deviations of quadruplicates are included, but SDs overlap with the symbols, as we found little or no variation. The linear regression curve of this amplified standard is indicated (r2 = 0.9978). (D) Single cells were retrotranscribed and preamplified (15 cycles) for Hprt-1. A second quantitative PCR was used for the evaluation of the relative expression level (CT) of Hprt-1 on each individual cell. Quadruplicates of amplification are shown. (E) Single individual cells were sorted, and treated for genomic DNA amplification (see Methods). The Gzma gene was next amplified in a seminested PCR (15 cycles of preamplification). PCR accumulation curves are shown for four individual cells. Data are from one of three independent experiments.











