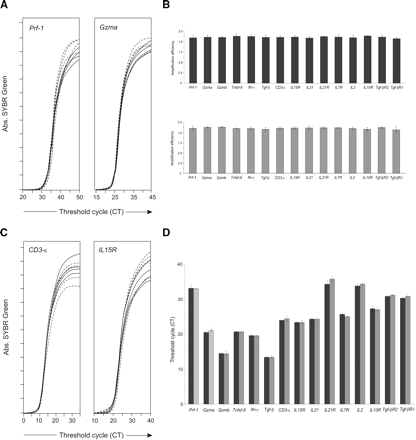
Efficiency and competition of PCR amplifications. (A,B) Aliquots of cDNA from mouse IEL were amplified separately for each gene and each type of PCR reaction to determine PCR efficiency. (A) Quadruplicate amplification slopes for Prf-1, Gzma. Set of primers from the first PCR (solid lines) the second PCR (dashed lines).The same tests were performed for all genes, giving the same results (B) Slope values from the quadruplicates were assessed on the exponential phase of the real-time amplification reaction, and PCR efficiencies were determined using LinRegPCR 7.0 software. Means ± SD of PCR efficiencies are shown for the first (upper panel) and second (bottom panel) PCRs. The significance of these differences was evaluated by ANOVA. Within each PCR, all primer combinations had the same efficiency. We also found no significant difference in the variance between the first and second PCR amplifications (ANOVA, P > 0.1). Data are from one of three independent experiments. (C,D) Competition: Aliquots of cDNA from IELs were amplified separately for each gene, or in multiplex in the first PCR round. Quadruplicates of these reactions were further amplified in a second real-time PCR. (C) Quadruplicates of amplification for Cd3-ϵ; IL15r, genes amplified in multiplex (solid line) or alone (dashed line). (D) Comparison of threshold cycle mean values (CT) obtained for each different gene amplified in multiplex (black bars) or separately (gray bars). No significant differences were observed between the two amplification conditions for each different gene (t-test, P > 0.1).











