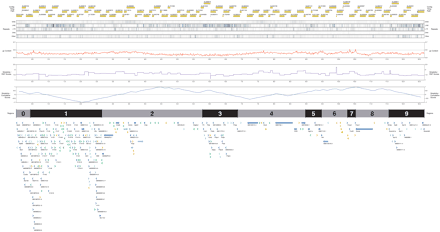
Pictorial representation of the annotation of the Del36H sequence contig. (Top to bottom) Yellow blocks represent the finished sequence of individual genomic clones forming the tiling path across this region; a black line or block at the start and/or end of a clone represents redundant overlapping sequence. Five tracks show in green the distribution of various types of repeats as defined by RepeatMasker. (Red) The distribution of the fraction of G and C nucleotides, between 0.3 (30%) and 0.7 (70%); (purple) the Relative Simplicity Factor scores; (blue) the cumulative signs plot of RSF scores. Alternating gray and black blocks indicate the segments of interest; odd-numbered segments contain gene clusters and evolutionary breakpoints, even-numbered segments lie between and around these. Arrows show the position of annotated genes, their relative transcriptional orientation, and their type; only genes of type “known” have their gene symbol shown. Dark blue: known genes; light blue: novel CDS genes; orange: novel transcript genes; green: pseudogenes; gray: putative genes.











