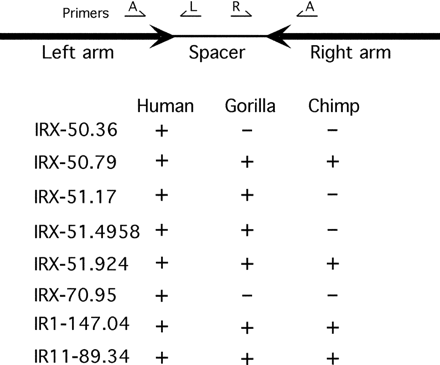
PCR analysis of internal arm/spacer boundary. For each IR indicated, a single primer (A) was designed to hybridize to both arms. Individual primers (L and R) were designed to hybridize to the spacer region. PCR amplification was performed using primer pairs A-L and A-R with human, gorilla, and chimpanzee genomic DNA. For each IR, a + indicates that PCR products were amplified using both primer pairs. All amplified IRs are >99% homologous (Table 1), except IRX-51.924, which is 97% homologous (Fig. 2A; Supplemental data S1 and S2). The PCR primers and DNA sequence of PCR products are shown in Supplemental data S3. The high homology between the arm and spacer of IRX-51.17 and IRX-51.924 (see Fig. 2B) does not allow for specific STSs, although two distinct sets of primers pairs were used (Supplemental data S3).











