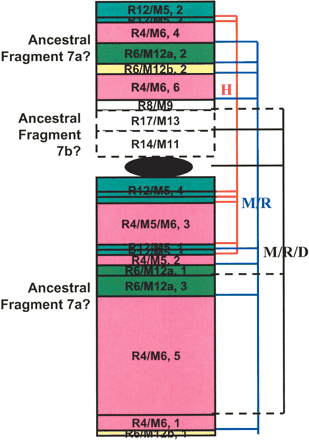
Intrachromosomal rearrangements in H7. H7p's first 32 Mb and the entire H7q are syntenic to R4(1-86 Mb)/M5(27-3)/M6(3-57), R12(9.7-27)/M5(143-128), R6(49-64)/M12(25-39)a, and R6(143-148)/M12(110-115)b (numbers in parentheses are in megabases). However, each of these continuous mouse/rat fragments broke and formed multiple syntenic blocks that scatter on both arms of H7. Blocks belonging to the same mouse/rat fragment are in the same color and numerically numbered on the basis of their orders in the fragment. For example, fragment R12/M5 split into four blocks with the second (labeled as “R12/M5, 2” in the plot) syntenic to the beginning of H7p and the other three to H7q. These portions of H7 are possibly derived from the large ancestral fragment H7a (blocks with solid lines). The rest of H7p, syntenic to R8(22-25Mb)/M9(22-25), R17(51.6-58)/M13(20-15), and R14(87-99)/M11(6.3-17), are possibly from the small ancestral fragment H7b (blocks with dashed lines). Block sizes are somewhat arbitrary and do not reflect the actual fragment length. Using dog as outgroup, among a total of 20 breakpoints shown, seven are due to human-specific intrachromosomal rearrangements (red lines on the right), eight are mouse/rat specific (blue lines), and five are shared by mouse/rat/dog (black lines, solid lines indicating where mouse/rat and dog break at the same site, dashed lines indicating where mouse/rat and dog break at slightly different [within 1-2 Mb] sites).











