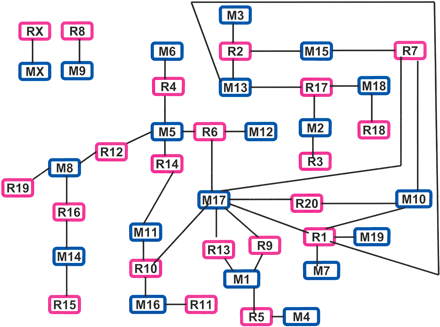
The mouse/rat synteny. Each block represents a mouse (blue) or rat (pink) chromosome (e.g., M1, mouse chromosome 1; R1, rat chromosome 1). A line was drawn between a mouse block and a rat block if synteny was found between the two chromosomes. This plot better reveals the interactions between the chromosomes within a genome compared with other synteny plots used (Waterston et al. 2002; Kirkness et al. 2003; Rat Sequencing Project Consortium 2004) as demonstrated by the following. (1) Except for the M9/R8 and X chromosomes, the rest of the chromosomes have exchanged genetic materials with other chromosomes forming a complex synteny network. (2) Within the network, chromosomes also display varying degrees of complexity in synteny. For instance, mouse chromosomes M3, M4, M6, M7, M12, and M19 are syntenic only to a single rat chromosome. The same is true for R3, R11, R15, R18, and R19. The remaining chromosomes are syntenic to 2 to 3 chromosomes from the other species, except for R1, M5, and M17 with synteny to 5, 4, and 7 chromosomes, respectively. (3) M17 has the most complex synteny. Compared with its rat homologs, M17 has additionally rearranged with M10 and M1 multiple times, as well as with M5, M11, and M16 at least once. Nearly every mouse/rat syntenic block in M10 and M1 has rearranged with M17, indicating subsequent fusions of these blocks forming the two chromosomes. (4) Chromosomes within the network seem to group together on the basis of the conservation and rearrangement between the two species. For instance, chromosomes located at the left (M8/M14 vs. R19/R16/R15), where rat is more rearranged, differ from those at the bottom, right corner (M17/M1/M4 vs. R9/R13/R5), where mouse is more rearranged. Similarly, whereas a large variation was found between M17/M10/M7/M19 and R7/R20/R1 at the right, a great conservation was identified between M3/M13/M15/M2/M18 and R2/R17/R3/R18/R7 at the top.











