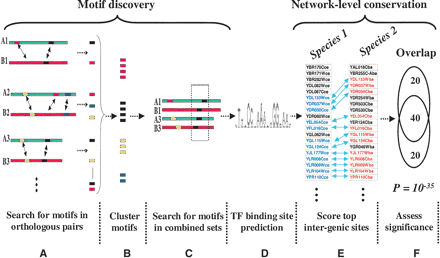
Schematic representation of the algorithm. (A) Upstream sequences from orthologous pairs of genes are searched to identify motifs using Gibbs-sampling. (B) Motif predictions are pooled and clustered by similarity. (C) The pairs of upstream sequences which yielded similar motifs (within a motif cluster) are combined and searched again for motifs using a second round of Gibbs-sampling. (D) A large number of motif predictions which need to be pruned. (E) To test for network-level conservation, the genes containing the top intergenic (5′ upstream) matches to each motif are identified in the two species. (F) The statistical significance of overlap between the two sets of genes is determined using the hypergeometric distribution.











