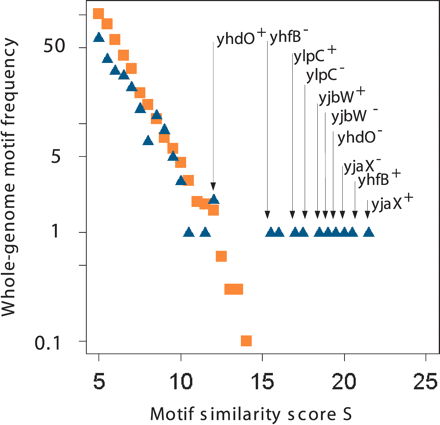
Whole-genome transcription-factor-binding site specificity. The position weight matrix of the TFBS motifs (see Fig. 2) has been used to search for similar motifs in the Bacillus subtilis whole-genome sequence. The plot displays the similarity score distribution (blue triangles), together with a score distribution derived from a random genome model (orange squares). Whereas the random hits obey a clear exponentially decaying distribution function even for higher scores, the B. subtilis genome-based results bend off at ∼S > 10. The genome-based results appear to be systematically underrepresented for lower scores, indicating a high genome-wide specificity of the identified TFBS motif. The arrows indicate the genes located downstream of the high scoring hits. Note that because of the palindromic nature of the motif, each gene appears twice, once for each DNA strand; (superscript +) orientation in transcription direction; (superscript -) opposite direction.











