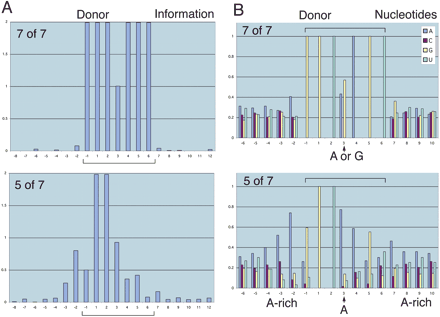
Partial-canonical splice sites reveal alternative information content. (A) The graphs show the information profiles for two sets of introns with lengths between 1024 and 4095 nt—those that match the donor consensus sequence GGU(A/G)AGU at D - 1 through D + 6 in all seven positions (7-of-7; 241 introns) compared with those that match at five positions (5-of-7; 123 introns). The 5-of-7 set had extra information outside of D - 1 through D + 6. The average standard deviation at a nucleotide position (-32 to 32) is 0.015 (7-of-7); 0.044 (5-of-7). (B) The graphs of nucleotide counts for the 5-of-7 set in A reveal a preference for A at D + 3 that would base-pair most effectively with U in U1 snRNA (Fig. 1A). The 5-of-7 set also had enriched A content at D - 3 through D - 1, which would enhance base-pairing with U5 snRNA (Fig. 1A). The standard deviation at each nucleotide position is <0.033 (7-of-7); <0.12 (5-of-7).











