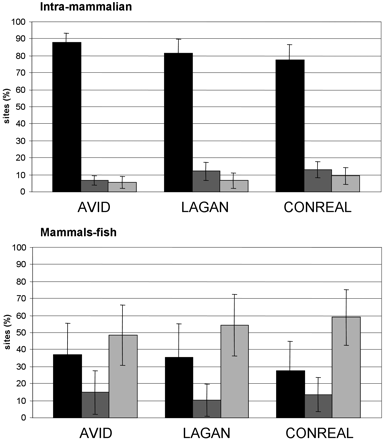
Intersection of the total number of conserved sites found by different approaches in intramammalian (top) and mammal–fish (bottom) gene pairs from the reference set. The percentage of sites found by all three methods is shown in black, sites confirmed by one additional method are dark gray, and method-specific fraction of sites is light gray. Error bars, 95% confidence intervals. The analysis parameters are 75% PWM threshold, 50% homology threshold, and 5-bp flank length.











