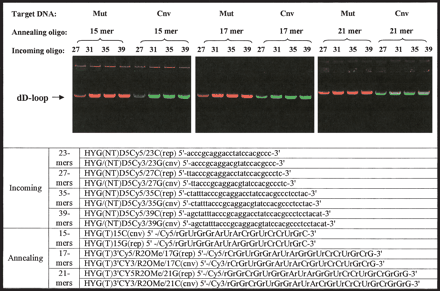
Size optimization of mismatch discrimination using labeled annealing oligonucleotides. The dependence of mismatch discrimination on the length of incoming and labeled annealing oligonucleotide combinations was assessed. Either linear 8-kb pAURHyg(wt)eGFP or pAURHyg(cnv)eGFP plasmids were used separately as target DNA as indicated. Mixed unlabeled incoming oligonucleotides (exactly matching mt and cnv targets, which differ by a single nucleotide) and mixed labeled 2′-O-MeRNA annealing oligonucleotides (exactly matching mt [Cy3, red] and cnv [Cy5, green] targets, which differ by a single nucleotide) were used. The ability to discriminate mismatches is strongly dependent on the size of both the incoming and annealing oligonucleotides. Incoming oligonucleotides 27, 31, 35, and 39 nt in length were tested as indicated. Annealing oligonucleotides 15, 17, and 21 nt in length were tested as indicated. Red bands indicate wt sequence (Cy5), green bands indicate cnv sequence (Cy3), and yellow indicates mixed signals.











