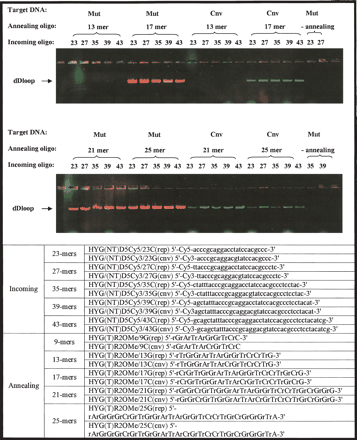
Size optimization of mismatch discrimination using labeled incoming oligonucleotides. The fluorescent gel picture depicts size optimization of combinations of the incoming oligonucleotides and the annealing oligonucleotides that accurately and reproducibly detect a particular gene variant. Mixed incoming oligonucleotides (exactly matching mt and cnv targets, which differ by a single nucleotide) and mixed annealing oligonucleotides (exactly matching mt and cnv targets, which differ by a single nucleotide) were used to form dD-loop complexes in either pAURHyg(wt)eGFP or pAURHyg(wt)eGFP linear plasmids as indicated. Incoming oligonucleotides are labeled with Cy5 (wt) and Cy3 (cnv), and 2′-O-MeRNA annealing oligonucleotides are unlabeled. Incoming oligonucleotides 23, 25 27 35, 39, and 43 nt in length were tested as indicated. Annealing oligonucleotides 13, 17, 21, and 25 nt in length were tested as indicated. Red bands indicate wt sequence (Cy5), green bands indicate cnv sequence (Cy3), and yellow indicates mixed signals.











