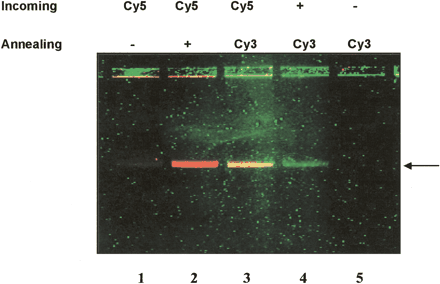
Concurrent detection of incoming and annealing oligonucleotides. The fluorescent gel picture shows the formation of a dD-loop in a linear 8-kb plasmid target with concurrent detection of differentially labeled incoming and annealing oligonucleotides. The incoming oligonucleotide is either unlabeled (no color) or Cy5-labeled (red). The incoming oligonucleotide is either unlabeled (no color) or Cy3-labeled (green) as indicated. (Lane 1) A Cy5-labeled incoming is unable to form a stable single D-Loop when no annealing oligonucleotide is added (no band). (Lane 2) Stabilization by adding a Cy5-labeled incoming and unlabeled annealing oligonucleotide in which the Cy5-labeled dD-loop is visualized as a red band. (Lane 3) Stabilization by adding a Cy5-labeled incoming and a Cy3-labeled annealing oligonucleotide in which both the incoming and annealing labels are colocalized and visualized as a yellow band combining red (Cy5) and green (Cy3) labels. The green band in lane 4 is created using an unlabeled incoming oligonucleotide and a stabilizing Cy3-labeled annealing oligonucleotide. (Lane 5) RecA is unable to form a stable single D-loop with a Cy3-labeled annealing oligonucleotide when no DNA incoming oligonucleotide is added (no band). The arrow indicates the position of the double D-loop as detected by fluorescence.











