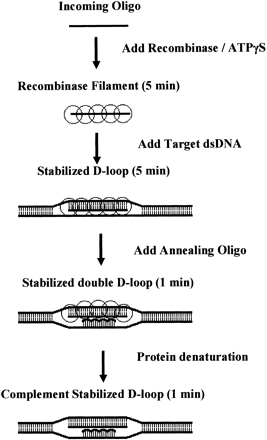
Schematic of standard double D-Loop formation assay. (1) A DNA “incoming” oligonucleotide is coated with RecA in the presence of a nonhydrolyzable ATP analog, ATPγS. (2) The preformed nucleofilament is added tothe duplex target substrate, and RecA catalyzes the sequence-dependent pairing and strand invasion of the duplex target, displacing one strand of the Watson-Crick base duplex to create a D-loop intermediate. (3) The D-loop intermediate is stabilized upon the addition of a second modified “annealing” oligonucleotide that is complementary to the displaced strand. (4) The resulting complement-stabilized double D-loop is analyzed after denaturing and dissociating RecA by adding SDS.











