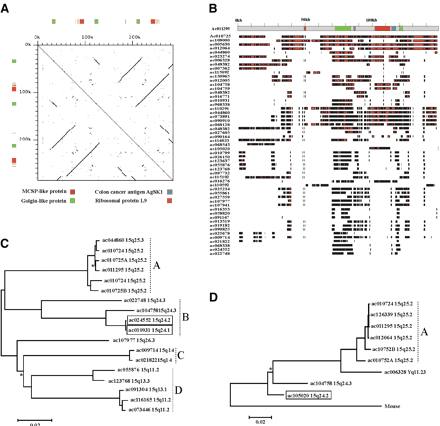
Analysis of 15q24-26 duplicons. (A) Dot matrix self-comparison of 15q25.2 duplicon (accession nos. AC011295 and AC010724; position 78744–79029 kb). A 30-bp perfect match was required to generate a dot, and a 30-bp offset was used. The position of gene-related sequences are indicated by colored boxes. The GLP and MCSP sequences span exons and introns of the functional genes, but do not retain the full coding sequences. The sequences share 81%–89% identity to the functional GLP gene and 82%–94% identity to the functional MCSP gene. The position of two unspliced transcripts that were highly similar to the region are also shown. BC004206 shares 100% identity to the sequence and is related to ribosomal protein L9. This is a retroposed copy of exons 2–8 of the true gene from chromosome 4. AF316855, an unspliced mRNA for colon cancer antigen AgSK1, shares 99.1% identity to the sequence but with a 61-bp insertion/deletion. We have also classed this sequence as a pseudogene. (B) Physical overlap of top 55 high-scoring BLAST hits with AC011295 (1–149 kb of A). High identity to the RepeatMasked test sequence is shown by black lines. The position of gene-related sequences (from A) and the scale in kb are shown. (C) Maximum likelihood tree of GLP sequences identified in (B). Alignment was constructed using sequences related to nt 71235–73570 of AC011295 which are within the GLP region but noncoding. Only sequences integrated into the working draft and the 15q24-26 map (Fig. 2) are included (see Methods). This tree has been arbitrarily rooted along the midpoint for ease of presentation, so ancestor-descendent relationships cannot be inferred from the topology. All nodes are supported by >95% bootstrap values with three exceptions, which are indicated with an asterisk. The sequences predicted to represent functional GLP genes (intact full-length ORFs and EST support, see Fig. 3) map to 15q24 and are boxed (AC010931, ac024552). All other GLP sequences are truncated with the exception of AC019294, which is full-length but contains multiple frame shift mutations. (D) Maximum likelihood tree of chondroitin sequences identified in (B). Alignment was constructed using exons 2 and 3 minus the intervening intronic sequence. Only sequences integrated into the working draft or the 15q24-26 map (Fig. 2) are included (see Methods). The mouse MCSP mRNA sequence was used as an outgroup. All nodes are supported by >95% bootstrap values with one exception, which is indicated with an asterisk. A trichotomy containing ac010724A, ac126339, and ac011295 is not clearly visible due to the extremely high sequence identities (∼99.9%) between these sequences and ac012064. The sequence predicted to represent the functional MCSP gene (intact full-length ORF with EST support, data not shown) maps to 15q24.2 and is boxed (AC105020). All other MCSP sequences are truncated relative to this sequence. A tree constructed using intronic sequences from the same loci (excluding mouse) gave results consistent with the topology shown (data not shown).











