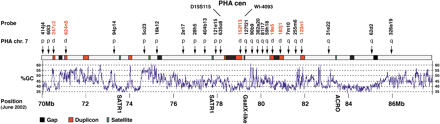
Sequence features within 15q24-26, and FISH results on PHA7. Human BAC clones used to probe PHA metaphases are indicated, together with the result of the hybridization. p, p arm only; q, q arm only; d, p and q arm, flanking the centromere (as in Fig. 1c). BACs RP11-152f13, RP11-182j1, and RP11-123n1 gave telomeric hits of lower intensity on PHA7p chromosomes. Red boxes indicate regions duplicated on normal human chromosome 15, identified in silico and confirmed by FISH. Green boxes show the location of satellite sequences; from left to right their sizes are 3675, 3019, 334, and 254 bp. Black boxes show the location of gaps between sequence contigs. The distances spanning the gaps at the ancient and neocentromere are a maximum of 2.5 cR (ancient cen), 0.1 cR (Case 1) and 1.2 cR(Case 2) as defined by the GM99 radiation hybrid map (http://corba.ebi.ac.uk/RHdb/species/HUMAN/gm99.html). Arrows denote the location of BACs used for FISH on the HSA15 contigs. The scale refers to the June 2002 human draft sequence, and the position of the ancestral centromere (PHA cen) is indicated.











