Table 2.
Folding Potential of Coding Regions From Diverse Organisms
|
Empire |
Species |
No. genes |
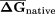 |
EFP |
Wilcoxon t-test |
%G + C |
|---|---|---|---|---|---|---|
| Archaea | Thermoplasma volcanium | 1525 | -143.2 | +1.0 | NS | 43.14 |
| Methanobacterium thermoautotropicum | 1873 | -204.0 | +4.0* | E-12 | 52.27 | |
| Sulfolobus solfataricus | 2977 | -121.8 | -0.2 | NS | 39.01 | |
| Sulfolobus tokodaii | 2826 | -110.6 | -0.2 | NS | 36.49 | |
| Pyrococcus abyssi | 1769 | -165.4 | +1.4 | NS | 47.22 | |
| Aeropyrum pernix | 1840 | -244.6 | +0.2 | NS | 58.86 | |
| Archaeoglobus fulgidus | 2407 | -187.6 | -0.6 | NS | 51.17 | |
| Halobacterium sp. NRC-1 | 2058 | -328.4 | +5.2* | <E-16 | 69.43 | |
| Eubacteria | Escherichia coli | 4290 | -218.6 | +4.8* | <E-16 | 52.90 |
| Salmonella typhi | 4600 | -227.6 | +6.0* | <E-16 | 54.30 | |
| Bacillus subtilis | 4100 | -165.6 | +2.8* | E-13 | 45.92 | |
| Mycoplasma pneumoniae | 689 | -147.6 | +6.6* | <E-16 | 42.58 | |
| Vibrio cholerae | 2752 | -193.2 | +4.4* | <E-16 | 50.13 | |
| Aquifex aeolicus | 1522 | -152.2 | +1.4 | NS | 45.52 | |
| Borellia burgdorferi | 850 | -98.4 | +1.6 | NS | 31.47 | |
| Thermotoga maritima | 1846 | -173.4 | +2.2* | E-05 | 48.33 | |
| Haemophilus influenzae | 1709 | -142.0 | +2.2* | E-05 | 40.79 | |
| Synechocystis PCC6803 | 3169 | -185.4 | -1.8* | E-07 | 50.35 | |
| Treponema pallidum | 1031 | -219.0 | +2.8* | E-03 | 54.85 | |
| Clostridium acetobutylicum | 3672 | -106.0 | +1.8* | <E-16 | 34.05 | |
| Rickettsia prowazekii | 834 | -102.6 | +1.2 | NS | 32.75 | |
| Helicobacter pylori | 1491 | -139.2 | -0.6 | NS | 41.96 | |
| Eukaryota | Plasmodium falciparum | 422 | -70.8 | +0.8 | NS | 26.60 |
| Saccharomyces cerevisiae | 6314 | -129.0 | +1.2* | <E-16 | 42.20 | |
| Arabidopsis thaliana | 2676 | -162.2 | -0.1 | NS | 47.80 | |
| Caenorhabditis elegans | 762 | -149.0 | -2.8* | E-07 | 46.40 | |
| Homo sapiens | 1855 | -207.8 | -0.8 | NS | 54.10 | |
|
|
Drosophila melanogaster
|
1956
|
-210.2
|
+0.01
|
NS
|
55.90
|
-
All sequences were randomized using the Dicodon Shuffle program. EFP as defined in Methods. (NS) Not significant.
-
↵* Statistically significant bias in favor (positive EFP) or against (negative EFP) RNA structure.











