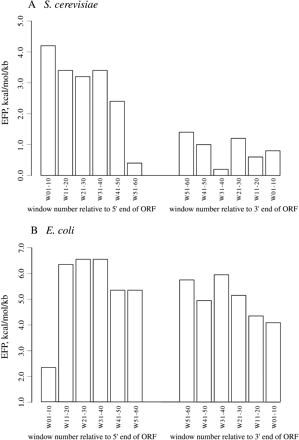
Distribution of EFP along coding regions in (A) E. coli and (B) S. cerevisiae. For native and shuffled mRNAs, six subsets of 50-bp sequence windows from each end of the coding region (corresponding
to the overlapping windows 1–10, 11–20, 21–30, 31–40, 41–50, 51–60 relative to the 5′ or 3′ end) were folded, average folding
free energies were calculated for native (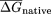 ) and CodonShuffled (
) and CodonShuffled (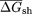 ) sequences, and the
) sequences, and the 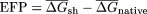 was determined for each segment. Because the step size between successive windows is 10 bp, each bin corresponds to 140
bases of sequence.
was determined for each segment. Because the step size between successive windows is 10 bp, each bin corresponds to 140
bases of sequence.











