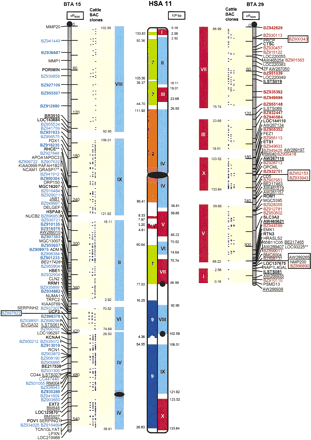
High-resolution cattle- and mouse-on-human comparative map of HSA11 created with current RH maps of BTA15 and BTA29 and 84 new cattle BESs. Comparative coverage on the current cattle RH5000 map is shown in solid red or blue blocks on the right half of the HSA11 drawing, with comparative segments indicated in Roman numerals. Mouse-on-human segments and mouse chromosome number are shown within the left half of the HSA11 drawing, with the sequence coordinates of the corresponding positions in the mouse genome indicated to the left. The distribution of BACs predicted to conserved segments of BTA15 and BTA29 is shown between the RH maps and human chromosome segments. Mapped conserved BAC ends (CASTS) increased the total comparative coverage and enabled merging of linkage groups for both cattle chromosomes. The CASTS are identified by GenBank accession number and are colored blue or red on chromosome maps. The human centromere is indicated by black ovals, and the 3.37-Mbp pericentric region on HSA11 with no BLAST hits is shaded. Black and gray circles indicate the location of centromeres on cattle and mouse chromosomes, respectively. Placement of markers appearing in boxes is defined by two-point linkage, because their order could not be determined with certainty. Framework markers are in bold text. Underlined markers are those with no hit against the human genome (microsatellites, and novel ESTs). Markers with two asterisks (**) are not predicted by COMPASS to be in the observed segment on the basis of the available comparative map. Comparative maps were drawn using NCBI Build 29 (April 2002) of the human genome.











