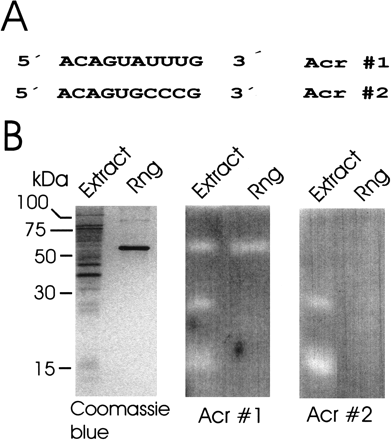
In-gel discrimination of a sequence-specific nuclease activity using a pair of oligonucleotide substrates. The sequences of the two substrates (Acr#1 and Acr#2) that were covalently linked separately to gel matrix are shown in A. Aliquots of an extract of E. coli BL21(DE3) and purified recombinant RNase G (Rng) were run side by side in three SDS polyacrylamide gels shown in B. The gel on the left did not contain substrate and was stained with Coomassie blue to indicate the position of the 58-kD Rng polypeptide. The gels in the middle and on the right contained Acr#1 and Acr#2, respectively, and were activity stained as described for RNase A in Fig. 1.











