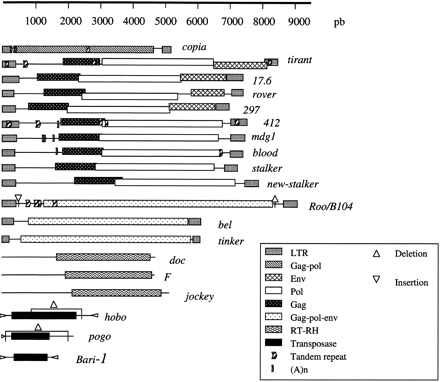
Structures of the sequences of the canonical transposable elements. Structures of the reference elements zam, doc2, and doc3 are not represented because there were no copies of zam in the sequenced genome and because doc2 and doc3 were very similar to the canonical doc element. The deletions indicated for the hobo and pogo elements correspond to the internal deletions that gave rise to the regulatory elements of 1406 bp for hobo and 806 bp for pogo. The deletion and insertion indicated for the roo/B104 element correspond to the deletion and insertion of 12 bp found in several copies. The tandem repeats and the (A)n indicated on the elements correspond to repeats for which variants in the number of occurrences were found in some copies in comparison to the canonical element.











