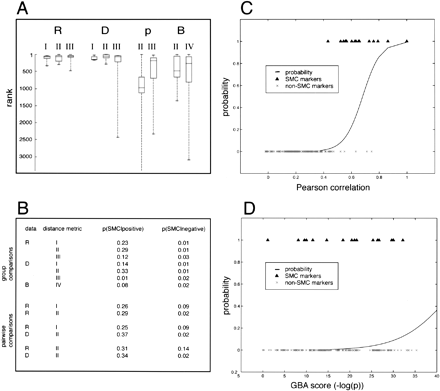
Systematic evaluation of alternative data types and distance metrics with respect to prediction of SMC-specific genes. R, transformed data; D, raw data; p, frequency data; B, binary data. Roman numerals indicate different distance metrics. I, Permutation; II, Pearson's correlation; III, covariance; IV, GBA (Walker et al. 1999). Euclidean distance was also evaluated, giving nonspecific results (not shown). For full definition of terms, see Methods. (A) Nearest neighbor searches were performed on all genes with at least five ESTs in UniGene (n = 29,812). Genes were ranked according to their profile similarity to SM-MHC. Ranks for 10 SMC markers are displayed on the Y-axis as box blots. Boxes, Central 20th to 80th percentiles; whiskers, the full range of observations. (B) Alternative methods (combinations of data type and distance metric) were evaluated with a logistic regression model. This model was used to compute probabilities for genes to be SMC markers based on their profile distance to SM-MHC. P(SMC|positive) denotes the model average probabilities for positive controls to be SMC markers, and (SMC|negative) denotes the corresponding probability for negative controls. Models were first compared group-wise using the full test set. This identified three preferred models (R-I, R-II, and D-II). These three models were then compared in a pair-wise fashion. To avoid bias, subsets of the reference gene set were used (see Methods). The preferred method was data = D, distance = II (Pearson's correlation), with expectations 0.37 for the markers and 0.02 for the nonmarkers. (C, D) Logistic regression curves for raw data/Pearson's correlation (C) and GBA (D). Triangles, Known SMC markers (probability = 1); crosses, genes with other expression pattern (probability = 0); curve, estimated relationship between a gene's correlation against SM-MHC and its probability to be an SMC marker.











