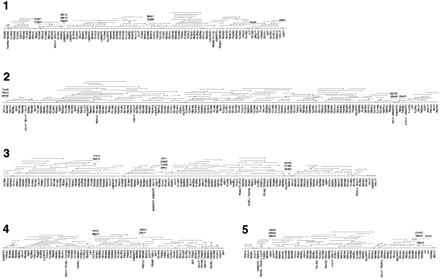
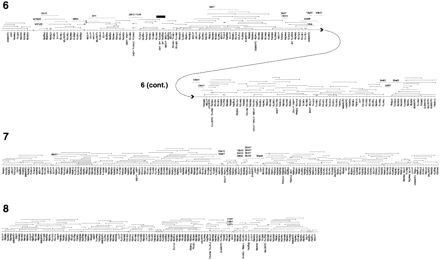
Genome-wide HAPPily anchored physical map. For clarity, markers are shown equally spaced. For each chromosome, HAPPY marker names are given below the map; names beginning `cr' and `CpG' indicate markers taken from Piper et al. (1998a) and from database sequences, respectively. Other names represent PAC end-sequences in condensed form; `q' = SP6 end, `p' = T7 end; for example, 06f03q is the SP6 end of the PAC clone pica_0006_f03. PAC clones are shown above each chromosome; the clone end represented by the mapped marker is indicated by a triangle; a short vertical line at the opposite end of a clone indicates that it ends within the interval shown; lack of this vertical line indicates that the clone may extend further than shown. Additional PAC or BAC clones identified during preliminary gap closure are indicated by dashed lines with the condensed clone name above them (names prefixed with `b' are BAC clones). Physical gaps are indicated by paired diagonal lines; the order of segments between these gaps is determined by the HAPPY data; the orientation of some of the smallest segments is not rigorously determined. Chromosome 6 is depicted in two parts; clone b3b01 is shown on both parts to illustrate the overlap. Solid triangles indicate deletions in clones; the solid black rectangle indicates a region of chromosome 6 which was sequenced by PCR amplification based on the corresponding part of the Moredun isolate genome (see text).











