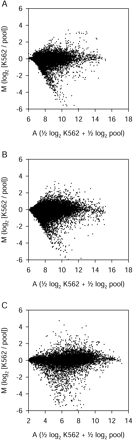
Differential expression and signal intensity measurements for all three array types. (A) Version 1 spotted long oligonucleotide arrays (means from six replicate two-color hybridizations). (B) Version 2 spotted long oligonucleotide arrays (four replicate two-color hybridizations). (C) In situ-synthesized 25-mer arrays (two replicate K562 sample single-color hybridizations and three replicate pool sample single-color hybridizations). Each point represents data from a single long oligonucleotide probe (A, B) or 25-mer probe set (C). M is a measure of differential gene expression (log2 [K562 intensity / pool intensity]). A is a measure of signal intensity (0.5 log2 K562 intensity + 0.5 log2 pool intensity).











