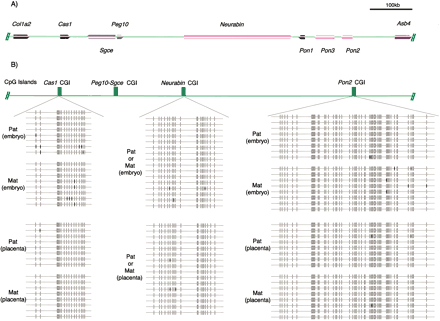
Genomic structure of the Col1a2-Asb4 region in the mouse. (A) Physical map of the genes identified in a 1-Mb region in the Mus musculus WGS supercontig Mm6_WIFeb01_97 (GenBank accession no. NW_000272). The arrows show the direction of each transcription unit, and imprinted genes are indicated by color: red and pink indicate strong and weak preferential maternal expression, respectively, and blue indicates complete paternal expression. Genes showing no expression biases between parental alleles are shown in black. (B) CpG islands (CGIs) and their DNA methylation states in the domain. There are four CGIs (length over 300 bp) shown in green boxes in the Col1a2-Asb4 region, and the three nonmethylated CGIs determined by bisulfite sequencing of genomic DNA isolated from (B6 × JF1) F1 embryo and placenta (day 10) are shown. DNA polymorphisms were used to determine paternal and maternal alleles of Cas1 CGI and Pon2 CGI, and showed that both alleles were nonmethylated in this region. No available DNA polymorphisms were found in Neurabin CGI, but no methylated CpGs were observed in this region either. Each horizontal line indicates the sequence from a single clone. Each CpG dinucleotide is represented by an oval. White and black ovals indicate nonmethylated and methylated CpGs, respectively. Details of the differentially methylated region of the Peg10-Sgce CGI are shown in Fig. 2A.











