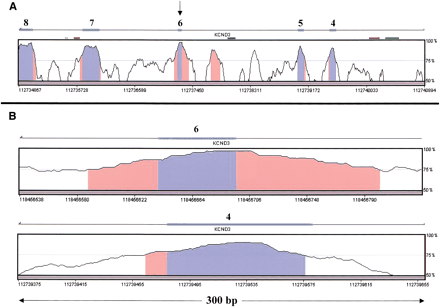

Human–mouse alignment of the KCND3 gene, corresponding to RefSeq NM_004980 (from VISTA browser, http://pipeline.lbl.gov/vistabrowser/). x-axis: The nucleotide coordinates on human chromosome 1, according to the assembly version of the human genome from June 2002. y-axis: The level of conservation between the human genome and the corresponding mouse genome, according to the MGSCv3 assembly version of the mouse genome. (A) Blue bars above the conservation area correspond to annotated exons 4–8 of KCND3. Blue areas within the conservation graph mark exons; orange areas mark conserved nonexonic sequences. The exon marked with an arrow (exon 6) is an alternatively spliced one; the others are constitutively spliced exons. (B) Enlarged view of the conservation graphs of the alternatively spliced exon (exon 6), and one of the constitutively spliced exons (exon 4) is presented to show the relative lengths of the conserved areas near the exons. (C) Human, mouse, and rat alignment of exon 6, as well as the 100 bases upstream and downstream of the exon. Exon sequence is bold; asterisks mark identity in all three organisms. Bold and underline mark the hexamer TGCATG, which previously showed the ability to regulate alternative splicing when found in introns downstream to alternatively spliced exons (Lim and Sharp 1998; Deguillien et al. 2001).











