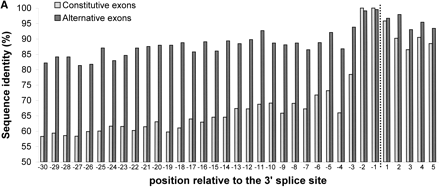
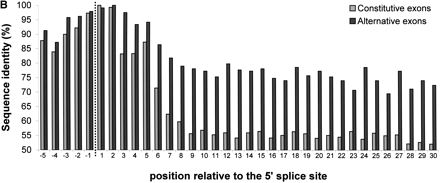
Per-position conservation near alternatively and constitutively spliced exons. Intronic regions near the splice site were aligned, using GAP (global alignment program) of the GCG package, and identity levels were calculated for each position as described in Methods. All 243 alternative exons and 1966 constitutive exons were used for this analysis. (A) Conservation near the 3′ splice site. Data for the last 30 nt of the intron and the first 5 nt of the exon are shown. Dashed line marks the border between the intron and the exon. (B) Conservation near the 5′ splice site. Data for the last 5 nt of the exon and the first 30 nt of the intron are shown.











