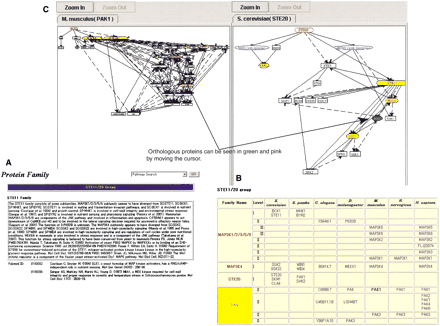
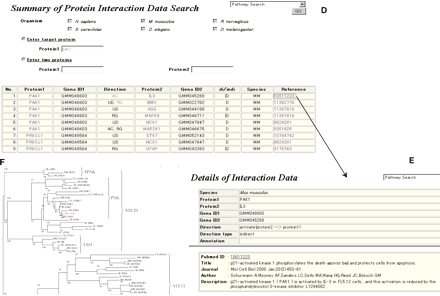
Interface for Kinase Pathway Database. (A) Comments about functional conservation in each family. (B)) Orthologous table. (C) Pathways viewer. Direct interaction and indirect interaction are respectively indicated by solid-lines and dotted liens. “Activate”, “inhibit”, and “regulate” are distinguished by the arrow shape. The blue nodes represent the clustered proteins, which have only single-step interactions. The yellow nodes represent kinases, which have ortholog tables. Moving the cursor, orthologous proteins are depicted in red (corresponding orthologous proteins) and green (by pointing with the cursor). (D) Protein interaction data. (E) References for protein interaction data (F) Phylogenetic tree. The red name is the query protein name.











