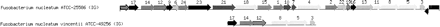
Deletion of the ethanolamine utilization operon in F. nucleatum sub spp vincentii. Between the ORFs for the dihydropteroate synthase (ORF12) and phosphoserine phosphatase (ORF 8) is the operon with 17 ORFs that encode enzymes for ethanolamine utilization found in F. nucleatum (Kapatral et al. 2002). The ORFs and their functional description are as follows: (17) GTP cyclohydrolase; (14) 2-Amino-4-hydroxy-6-hydroxymethyl dihydropteridine pyrophosphokinase/dihydroneopterin aldolase (fusion protein); (12) dihydropteroate synthase; (9) EutS; (6) EutP; (24) two-component response regulator; (23) sensor kinase; (21) EutA; (18) ethanolamine ammonia-lyase heavy chain; (15) ethanolamine ammonia-lyase light chain; (1) EutL; (10) EutM(potential frame shift); (7) EutM(potential frame shift); (4) EutE; (2) cobalamin adenosyltransferase; (22) hypothetical protein; (19) EutN; (16) hypothetical protein; (13) ethanolamine permease; (11) EutQ; (8) phosphoserine phosphatase; (5) butanol dehydrogenase; (3) thioredoxin.











