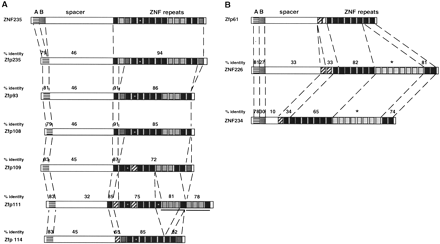
(A) Comparison of predicted proteins encoded by ZNF235, Zfp235, and five other mouse genes in Group IV. Entire proteins were aligned to maximize amino acid sequence identities; the order in which the genes are listed here is not meant to indicate a hypothesis of a linear series of duplication events. Boxes indicate KRAB-A and -B domains and ZNF repeats schematically. Diagonally-striped boxes are degenerate finger repeats. Some of the functional finger repeats are filled in shades of gray to help diagrammatically indicate chosen fingers that are present in most proteins but are absent in another. The sequence of Zfp111 contains evidence for at least one internal repeat-duplication event (underline) and possibly a remnant of a second (dot on suspected duplicated finger); three of the fingers duplicated in Zfp111 (light gray) are deleted in Zfp114. The numerical values over subregions of the predicted proteins indicate the amino acid sequence identity between ZNF235(top) and proposed homologous regions of each of the six mouse proteins, omitting the sections that are absent in either protein. (B) Comparison of the Zfp61, ZNF226, and ZNF234 proteins. Predicted complete proteins were aligned to maximize amino acid sequence identities. KRAB-A and -B domains and ZNF repeats are indicated by boxes (with diagonally striped boxes for degenerate fingers as above). Numerical values over subregions of the proteins indicate the amino acid sequence identity between Zfp61 and ZNF226 or Zfp61 and ZNF234; therefore there is no value for the block of fingers shared by the two human proteins but absent (probably due to a deletion event) in Zfp61 (*).











