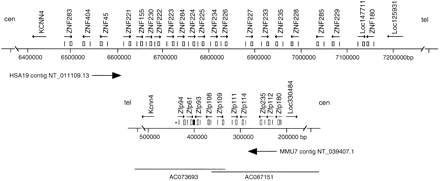
Maps of the homologous human and mouse ZNF gene family regions. Positions of KRAB-A- (vertical lines) and ZNF-encoding exons (boxes) that comprise gene models for 21 human and 10 mouse ZNF genes, as designated by arrows over the corresponding exons indicating transcriptional direction, are drawn above maps of relevant portions of HSA19q13.2 sequence contig NT_011109.13 (top) and Mmu7 contig NT_039407.1 (bottom). Locations of flanking markers KCNN4 and human LOC125931 (mouse LOC330484) are also shown. Numbers below each map correspond to nucleotide positions in the sequenced contig. The regions surrounding mouse and human gene clusters are inverted in telomeric-centromeric orientation due to an ancient chromosome rearrangement event, as indicated by symbols “tel” and “cen” above each map. To align homologous genes, we therefore display the reverse complement of the mouse contig sequence (as indicated by arrow below the mouse map). Conflicts between the NT_039407.1 sequence assembly and cDNA sequences were resolved by examining draft sequence from two overlapping BACs, the approximate extent of which is illustrated by lines drawn at the bottom of the mouse map. Associated numbers correspond to GenBank accession numbers for the BAC sequences. In addition to 10 complete genes, the mouse region contains an isolated ZNF-like segment without a significant ORF (filled box, at positions 407113–407607 at the 3′ end of Zfp61) and a single isolated KRAB-A sequence that is not associated with a known gene (positions 442889–443028 on NT_039407.1, indicated by an asterisk).











