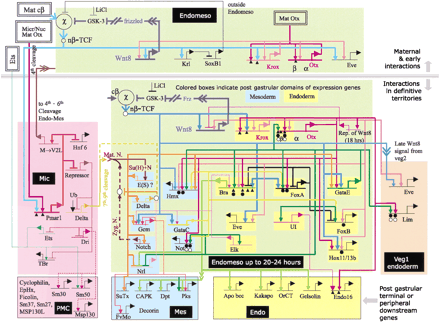
Hierarchical network of gene regulation involved in the specification of endoderm and mesoderm during early sea urchin development (Reprinted with permission from Davidson et al. 2002. Copyright 2002 American Association for the Advancement of Science.). Beginning with the detailed analysis of the cis-regulatory elements flanking a single gene involved in this process, endo16, as well as various trans-acting factors involved in the regulation of endo16, a network of genes and their levels of mRNA expression were studied. Regulatory interactions were determined by antisense oligo inhibition, injection of mRNAs into the embryo, and other perturbation methods. The network depicted in the diagram is based on modeling the various empirical parameters, including the experimental effects in embryos, using a computational environment on the basis of principles used to model mixed analog-digital circuits in the microelectronics industry. For details, see Brown et al. (2002). This envisioned network is but one example of how systems biology is conceptually impacting the field of gene expression. This figure also conveys how challenging it would be at the present time to relate the depicted gene regulatory circuits in relation to nuclear organization.











