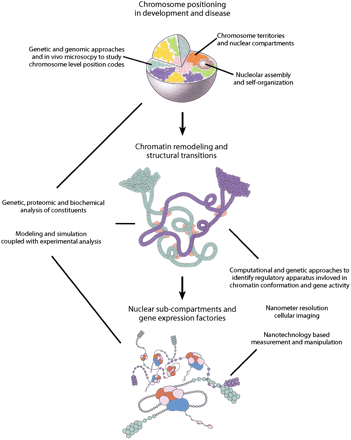
The information contained in the genomic DNA sequence is packaged within the nucleus. Multilevel hierarchical interactions are theorized to modulate the spatial arrangement and packaging of DNA in order to decode and process this information. These multilevel interactions are envisioned to exist on several scales to influence the relative position of chromosomes and the formation of the variety of supramolecular assemblies associated with dynamic nuclear compartments and chromatin domains. (Top) Individual chromosomes represented as different colored domains occupy definable territories within the interphase nuclear volume. Organizational principles that influence the location and potential nonrandom positioning of chromosomes relative to each other in the interphase nucleus are proposed to reflect an interplay between chromosome composition and sequence-based, protein-mediated associations with the nuclear envelope, nuclear pore complexes, and other chromosomes, compartments, and factors in the nuclear milieu. In the chromosome territory, DNA is likely to be organized in a variety of condensed and decondensed structural conformations. (Middle) Chromatin remodeling and scaffolding complexes (light red ovals) are proposed to generate domains within and between chromosome territories that contain extended chromatin fibers (green and blue loops). The extension of the chromatin fiber potentially provides an opportunity for further interactions between nuclear factors and loci leading to the assembly of various nuclear compartments. For example, positioning a gene cluster away from heterochromatin may favor the formation of a gene expression factory in which conditions are more permissive for RNA production and processing. Further remodeling of the chromatin provides transcriptional accessory proteins with increased access to the regulatory apparatus for individual or sets of genes. (Bottom) Transcriptional accessory proteins (red and purple clusters) interact with regulatory elements and the basal transcriptional machinery (light blue oval) to promote or repress the expression of a gene. Global as well as local modulations in chromosome organization and the packaging of DNA are needed to achieve cell-type specific profiles of gene expression.











