
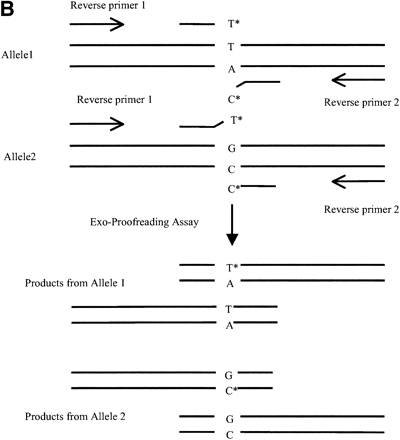
(A) Diagram of three-primer Exo-Proofreading assay. A primer is designed with its 3′ terminus aligned at the polymorphic base of a DNA target (T to C in this example) and labeled with a detectable tag on the 3′ nucleotide base. If the 3′ end of the primer is matched with the target (allele 1), the labeled nucleotide is retained and incorporated into a PCR product generated with a reverse primer. If the primer has a mismatch with the template (allele 2), the labeled nucleotide is removed by the proofreading activity of the polymerase and no tag is incorporated into the PCR product. (B) Diagram of four-pPrimer Exo-Proofreading assay. The four-primer assay is essentially the same as the three-primer assay except that the forward primers are interrogating the SNP from the plus and minus strands of DNA. An extra reverse primer is added, resulting in two PCR products.











