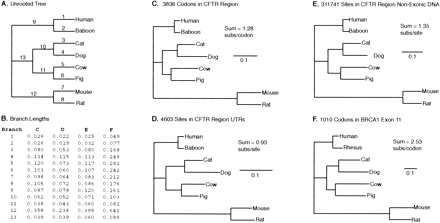
The tree relating the organisms of this study, and the number of substitutions per site in each branch of the tree as estimated from our four data sets. (A) The unrooted topology relating eight mammals from which CFTR region sequence data are available. Numbers on branches are identifiers that correspond to the rows inB. (B) Likelihood estimates of the number of substitutions per site or per codon in each of the tree's branches. Note that substitutions per codon represent a cumulative measure of the number of nucleotide substitutions for each codon, consisting of three nucleotide sites. (C–F) The four data sets. Unrooted trees with branch lengths drawn proportional to the number of substitutions per site or per codon as listed in B. Data set and the number of sites or codons is shown at top. (Sum) Total length of the tree. Scale bar, 0.1 substitutions per site or per codon.











