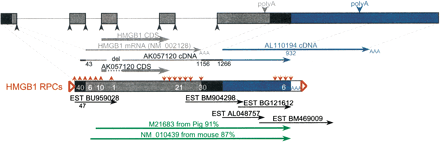
Transcripts of the human HMGB1 gene: alternative 3′ end, extended 5′ end. RPCs aligned directly to the HMGB1 genomic DNA (gray boxes) were found to have sequences extending upstream and downstream (black and blue boxes) relative to the HMGB1 RefSeq mRNA NM_002128. The middle structure is a schematic representing all RPCs derived from HMGB1, with gray structures corresponding to NM_002128 sequence, blue structures corresponding to a cDNA downstream from NM_002128 (AL110194), and black boxes are genomic sequences outside these cDNAs. The numbers within the boxes show how many RPCs start (with upward-pointing red arrows) or end (with downward-pointing red arrows) within this region. ESTs shown belong to UniGene cluster Hs.337757, and illustrate the alternative use of poly(A) signals (downward-pointing blue and gray arrowheads), as well as the existence of transcription upstream and downstream to NM_002128, also supported by a cDNA (AK057120). (Green arrows) Pig and mouse HMGB1 mRNAs, which also extend downstream of NM_002128. (Open triangles) TSDs; (AAA) poly(A) tracts; (upward-pointing black arrowheads) splice sites. The dotted lines in AK057120 depict a 70-bp deletion, and 3′ EST orientations are reversed.











