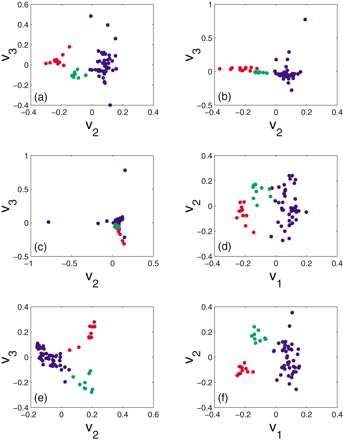
Lymphoma: Scatter plot of experimental conditions of the two best class partitioning eigenvectors vi,vj. The subscripts (i,j) of these eigenvectors indicate their corresponding singular values. CLL samples are denoted by red dots, DLCL by blue dots, and FL by green dots. (a) Bistochastization: the 2nd and 3rd eigenvectors of BBT . (b) Biclustering: the 2nd and 3rd eigenvectors ofR −1 AC −1 AT . (c) SVD: the 2nd and 3rd eigenvectors ofAAT . (d) Normalization and SVD: the 1st and 2nd eigenvectors of ĀĀ Twhere Ā is obtained by first dividing each column of A by its mean and then standardizing each row of the column normalized matrix. (e) Normalized cut algorithm: 2nd and 3rd eigenvectors of the row-stochastic matrix P. P is obtained by first creating a distance matrix S using Euclidean distance between the standardized columns of A, transforming it to an affinity matrix with zero diagonal elements and off diagonal elements defined asWij = exp(−αSij )/max(Sij ) and finally normalizing each row sum of the affinity matrix to one. (f) As in (c) but with an SVD analysis of the log interaction matrix K instead of A.











