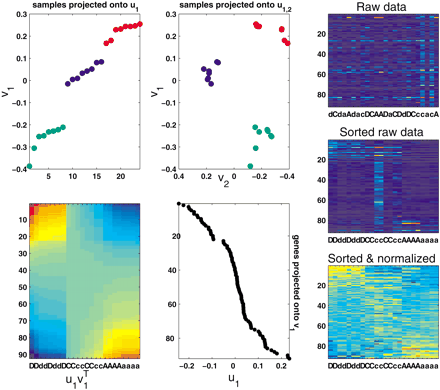
Optimal array partitioning obtained by the 1st singular vectors of the log-interaction matrix. The data consist of eight measurements of mRNA ratios for three pairs of cell types: (A,a) benign breast cells and the wild-type cells transfected with the CSF1R oncogene causing them to invade and metastasize; (C,c) cells transfected with a mutated oncogene causing an invasive phenotype and cells transfected with the wild-type oncogene; and (D,d) cells transfected with a mutated oncogene causing a metastatic phenotype and cells transfected with the wild-type oncogene. In this case we preselected differentially expressed genes such that for at least one pair of samples, the genes had a threefold ratio. The sorted eigen-gene v1 and eigen-arrayu1 have gaps indicating partitioning of patients and genes, respectively. As a result, the outer product matrix sort(u1 ) sort(v1 )T has a “soft” block structure. The block structure is hardly seen when the raw data are sorted but not normalized. However, it is more noticeable when the data are both sorted and normalized. Also shown are the conditions projected onto the first two partitioning eigenvectors u1 and u2. Obviously, using the extra dimension gives a clearer separation.











